Library Prep for The ENCORE Priming Project in Bermuda
Library Prep for The ENCORE Priming Project in Bermuda
Protocols used
This post details the info about the SwitchFree library prep steps to produce adequate libraries for RNA sequencing with Madracis decactis. I’m using the Zymo-Seq SwitchFree 3’ mRNA Library Kit. I followed Dr. Jill Ashey’s post here and Zymo’s protocol here.
I will be prepping samples from the ENCORE priming experiment conducted in Bermuda 2024.
The kit needs a minimum of 10 ng of total RNA or a maximum of 500 ng of total RNA, which is a large range. We will be using 12 ng of total RNA as input. Here’s a breakdown of input RNA volumes for each sample:
I will be doing 1 samples with the modifications below
Modifications
- Used 12 ng RNA input instead of 11 ng
- For the polyA R1 reagent, used 3uL of polyA R1 reagent + 2uL of DNase/RNase free water instead of 5 uL of polyA R1 reagent
Total RNA volume used was rounded to 3 decmal places and used to calulate the DNAse/RNAse Free Water used
| Sample ID | TS RNA (ng/uL) | RNA (uL) | DNAse/RNAse Free Water (uL) | Total starting volume (ul) | Primer |
|---|---|---|---|---|---|
| MD-5 | 38.2 | 0.314136 | 4.685 | 5.0 | 28 |
Zymo SwitchFree library prep workflow

Here I’m using UDI primers from the 96 well plate (CAT. D3096, LOT. 255118) provided with the kit.
Materials
- Zymo-Seq SwitchFree 3’ mRNA Library Kit
- PCR tubes
- Thermocycler
- Mini centrifuge
- Aluminum beads (to keep things on ice)
- Magnetic stand for PCR tubes
- Kit contents

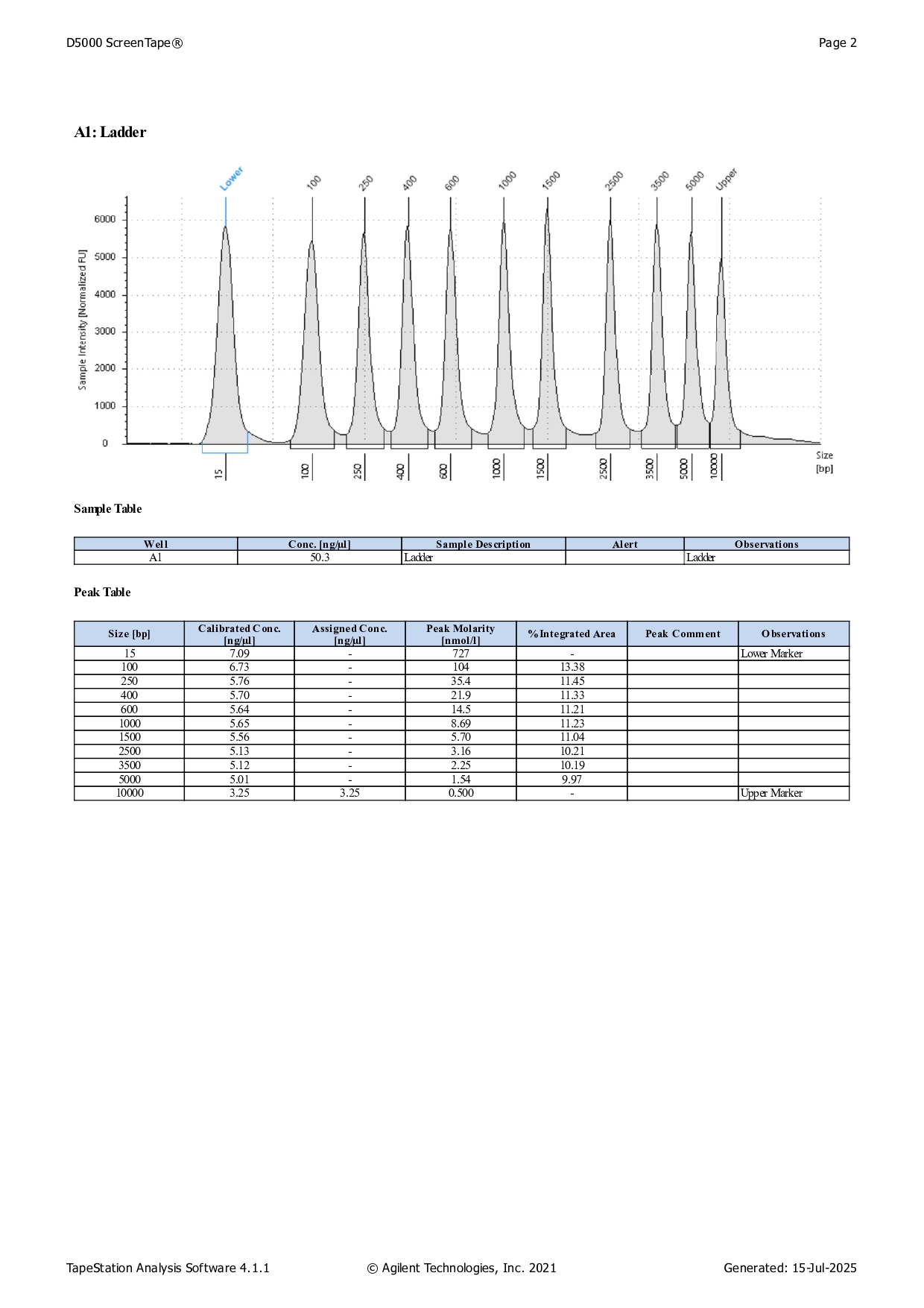
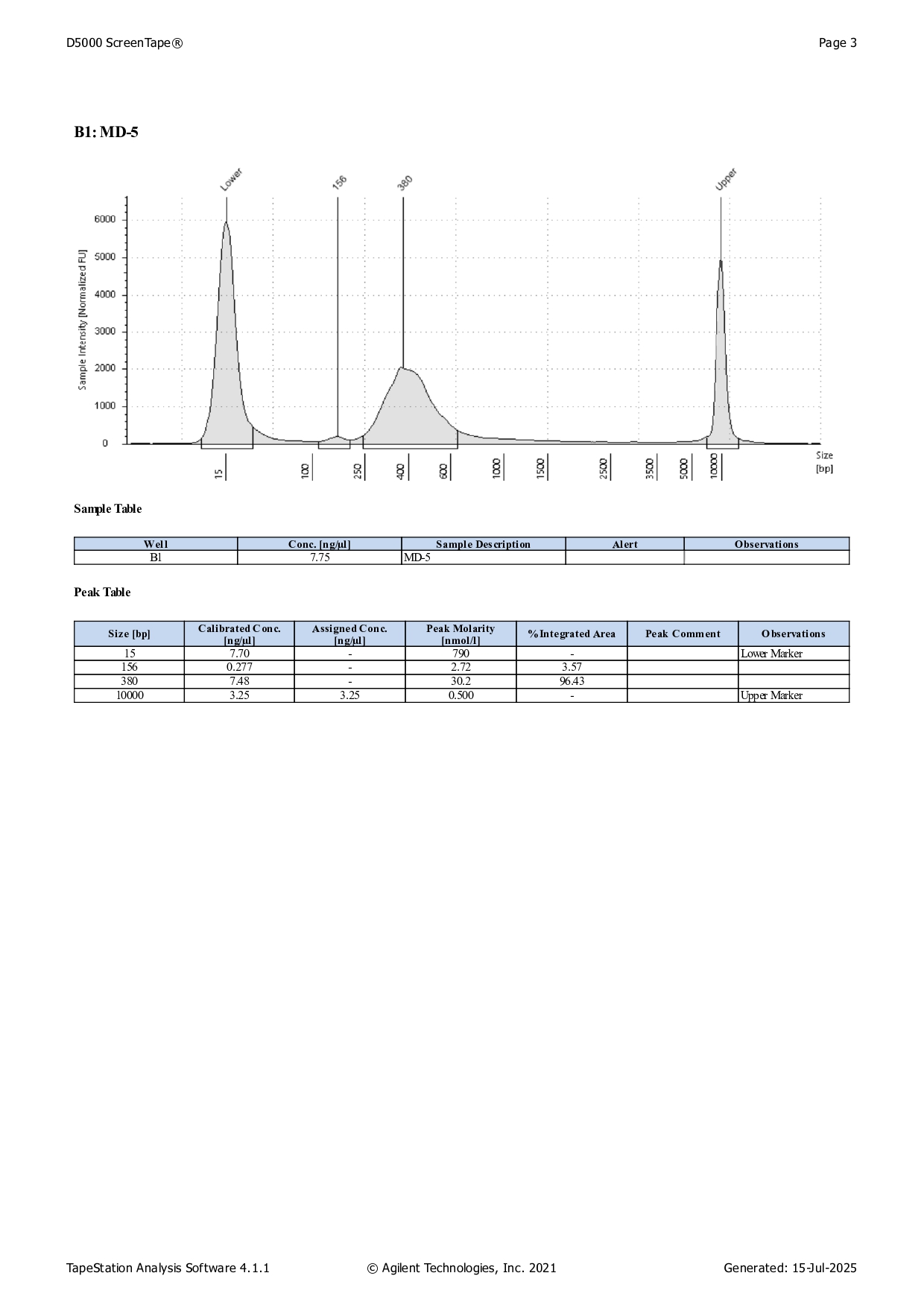
Qbit dsDNA HS results
See here for protocol.
| Sample ID | ug/uL #1 | ug/uL #2 | avg ug/uL |
|---|---|---|---|
| Standard #1 | 50.55 | ||
| Standard #2 | 24004.99 | ||
| MD-1-19 | 5.34 | 5.32 | 5.33 |
See the full report here.
Note: Samples MD-4-24 had many peaks and needs to be redone